7 Emulator diagnostics
A video presentation of this section can be found here.
The function validation_diagnostics can be used as in the deterministic case, to get three diagnostics for each emulated output.
We call validation_diagnostics passing it the mean emulators, the list of targets, the validation set:
vd <- validation_diagnostics(stoch_emulators$expectation, targets, all_valid, plt=TRUE, row=2)

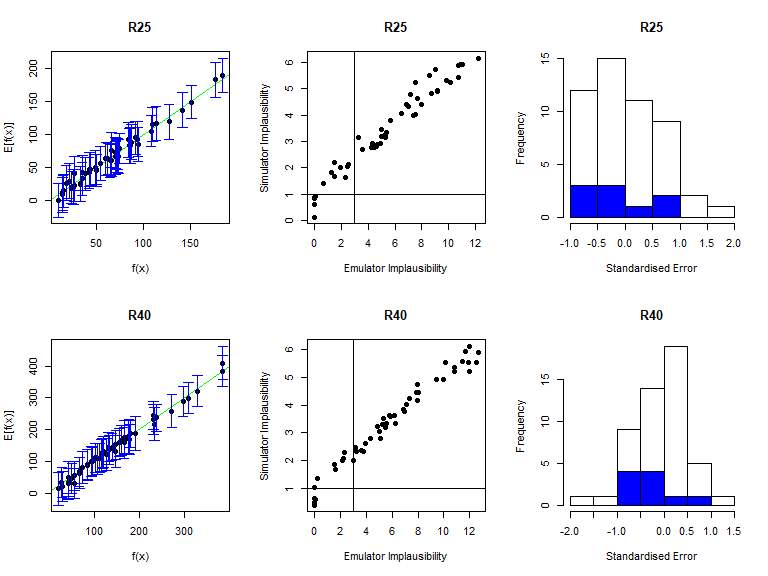
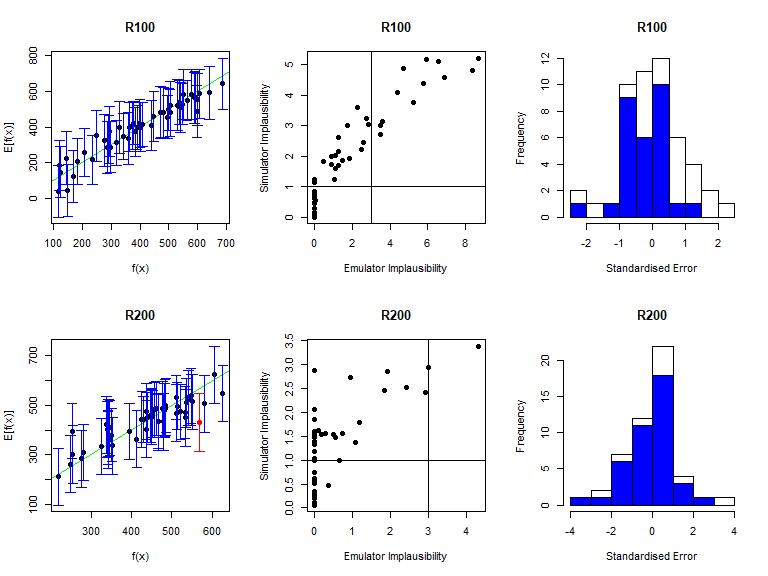
Note that by default validation_diagnostics groups the outputs in sets of three. Since here we have 8 outputs, we set the argument row to 2: this is not strictly necessary but improves the format of the grid of plots obtained. As in deterministic case, we can enlarge the \(\sigma\) values of the mean emulators to obtain more conservative emulators, if needed. Based on the diagnostics above, all emulators perform quite well, so we won’t modify any \(\sigma\) values here. In order to access a specific mean emulator, you can type ems$expectation$variable_name. For example, to double the \(\sigma\) of the mean emulator for \(I25\), you would type stoch_emulators$expectation$I25 <- stoch_emulators$expectation$I25$mult_sigma(2).
Note that the output vd of the validation_diagnostics function is a dataframe containing all parameter sets in the validation dataset that fail at least one of the three diagnostics. This dataframe can be used to automate the validation step of the history matching process. For example, one can consider the diagnostic check to be successful if vd contains at most \(5\%\) of all points in the validation dataset. In this way, the user is not required to manually inspect each validation diagnostic for each emulator at each wave of the process.
It is possible to automate the validation process. If we have a list of trained emulators ems, we can start with an iterative process to ensure that emulators do not produce misclassifications:
check if there are misclassifications for each emulator (middle column diagnostic in plot above) with the function
classification_diag;in case of misclassifications, increase the
sigma(say by 10%) and go to step 1.
The code below implements this approach:
for (j in 1:length(ems)) {
misclass <- nrow(classification_diag(ems[[j]], targets, validation, plt = FALSE))
while(misclass > 0) {
ems[[j]] <- ems[[j]]$mult_sigma(1.1)
misclass <- nrow(classification_diag(ems[[j]], targets, validation, plt = FALSE))
}
}The step above helps us ensure that our emulators are not overconfident and do not rule out parameter sets that give a match with the empirical data.
Once misclassifications have been eliminated, the second step is to ensure that the emulators’ predictions agree, within tolerance given by their uncertainty, with the simulator output. We can check how many validation points fail the first of the diagnostics (left column in plot above) with the function comparison_diag, and discard an emulator if it produces too many failures. The code below implements this, removing emulators for which more than 10% of validation points do not pass the first diagnostic:
bad.ems <- c()
for (j in 1:length(ems)) {
bad.model <- nrow(comparison_diag(ems[[j]], targets, validation, plt = FALSE))
if (bad.model > floor(nrow(validation)/10)) {
bad.ems <- c(bad.ems, j)
}
}
ems <- ems[!seq_along(ems) %in% bad.ems]Note that the code provided above gives just an example of how one can automate the validation process and can be adjusted to produce a more or less conservative approach. Furthermore, it does not check for more subtle aspects, e.g. whether an emulator systematically underestimates/overestimates the corresponding model output. For this reason, even though automation can be a useful tool (e.g. if we are running several calibration processes and/or have a long list of targets), we should not think of it as a full replacement for a careful, in-depth inspection from an expert.